Flying under the radar: Multi-drug-resistant bacteria hides among gut bacteria of asymptomatic human carriers
Scientists from A*STAR's Genome Institute of Singapore (GIS) have discovered how Carbapenemase-producing Enterobacteriaceae (CPE)—a multi-drug-resistant bacteria—hides among gut bacterial communities in asymptomatic human carriers. The study was published in Nature Microbiology.
Long-term colonization of the gut microbiome by CPE, a family of bacteria resistant to most antibiotics, is a growing area of public health concern as it can lead to community transmission and life-threatening CPE infections. Individuals may experience CPE bacteria colonization (e.g. E. coli or K. pneumoniae) through contact with contaminated surfaces or consumption of contaminated foods; prolonged consumption of antibiotics can also be a risk factor for colonization. Some individuals can develop CPE infections, and treatment often involves using several antibiotics of "last resort". In some severe cases, treatment failures lead to a high mortality rate (up to 50%) in hospitalized patients.
One place where scientists believe CPE hides is in the human gut, colonizing it asymptomatically and masked by the trillions of good bacteria there, waiting for the opportunity to cause infections or transmit to other individuals.
To study the effects of CPE colonization, and subsequent decolonisation on the gut microbiome, the team collaborated with Tan Tock Seng Hospital (TTSH) to examine the gut microbiome of CPE-positive subjects and their family members for one year.
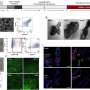
They tracked the gut bacteria by analyzing their metagenomic DNA signatures and discovered that CPE is not completely silent in the gut. The team found that CPE colonization leaves tell-tale marks by depleting key bacteria whose functions are linked to maintaining gut health by keeping inflammation in check.
The team also discovered that CPE colonization was dynamic, and could potentially lead to the exchange of genetic elements that confer antibiotic resistance across bacterial species.
By analyzing the whole genome sequences of CPE strains, they found mutations in key functional genes, and differences in antibiotic resistance profiles between highly similar Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae) strains that could explain why CPE is able to successfully hide in the gut. Rebalancing gut bacteria, particularly in terms of key species that modulate gut inflammation could provide an avenue for promoting CPE decolonisation, and preventing onward transmission of antibiotic resistance.
Dr. Niranjan Nagarajan, Group Leader of the Laboratory of Metagenomic Technologies and Microbial Systems and Associate Director of Genome Architecture at GIS, as well as corresponding author of the study, says that "antimicrobial resistance is a major public health challenge with a large impact on health care systems in Singapore. Our study sheds light on an important phenomenon that is of concern to infectious disease clinicians around the world."
"CPE are particularly worrisome in terms of antimicrobial resistance because they carry the resistance genes on mobile elements that can be exchanged between bacteria, thus rapidly creating superbugs out of otherwise harmless bacteria. They are like deadly assassins hiding in plain sight among gut bacteria. Yet, their presence does not go unnoticed by the gut bacterial crowd, and perhaps the key to eliminating them is to harness the power of the crowd."
Dr. Jonathan Teo Jin Yuan, Research Fellow at the Laboratory of Metagenomic Technologies and Microbial Systems in GIS, and joint first author of the study, says that "prior studies on CPE focused largely on the epidemiology and molecular aspects of the infection, but the natural history of CPE gut colonization has remained unexplored. Our study was unique as it followed up with CPE-colonized patients and their family members for a year, allowing us to track changes in their gut bacterial communities."
"We used cutting-edge metagenomic technologies to deeply probe the genetics of their gut bacteria, track the changes at the whole-genome level, and subsequently, analyzed how their gut bacterial communities evolved in relation to colonization by CPE."
Prof Patrick Tan, Executive Director of GIS, says that "antimicrobial resistance is often spoken of as the next pandemic threat that could overwhelm health care systems worldwide. Among the organisms of concern, CPE are close to the top of the list because of their ability to transmit antibiotics resistance and colonize the human gut, even in the absence of overt infection and antibiotic usage."
"By studying CPE gut colonization using advanced genomic tools, we were able to discover key insights that could help us reduce gut colonization, prevent the spread of these pathogens, and hence, safeguard our community and health care systems."
More information: Jonathan T. L. Kang et al, Long-term ecological and evolutionary dynamics in the gut microbiomes of carbapenemase-producing Enterobacteriaceae colonized subjects, Nature Microbiology (2022). DOI: 10.1038/s41564-022-01221-w
Journal information: Nature Microbiology